Research
Areas of Research: Protein post-translational modification (PTM) by ubiquitin (Ub) and ubiquitin-like proteins; Probes, inhibitors and assay platforms against deubiquitinases (DUB); Ubiquitin ligase, protein degradation and drug development through PROTACs; Diagnostics and therapeutics in neurological disorders such as Parkinson’s disease and cancer.
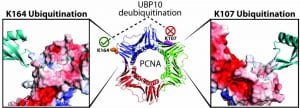
Protein ubiquitination through chemical approaches
We develop new methods for efficient protein mono- and poly-ubiquitination. Chemical ubiquitination circumvents the requirement for the ubiquitin cascade enzymes and can be readily generalized for modifying different target proteins. It also allows the generation of homogeneous ubiquitinated proteins with improved yields. Currently we are exploiting a diverse array of chemistries for site-specific protein ubiquitination. The chemically ubiquitinated proteins allow investigation of the molecular mechanism of biological processes that involves ubiquitination and deubiquitination, including DNA damage tolerance, mitophagy and innate immune response.
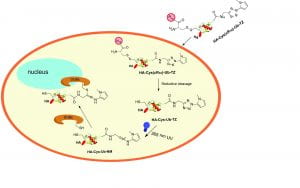
Ubiquitin-based deubiquitinase probes and assays
To study the linkage and target protein specificity of deubiquitinases, we have developed linkage-specific di- and poly-ubiquitin probes and cell-permeable DUB probes that allowed the interrogation of DUB specificity and function in both purified form and live cells. The ligation chemistry and linker molecules developed in our lab are applied to study ubiquitination of other proteins known to modified post-translationally by ubiquitin and ubiquitin-like proteins such as ISG15.
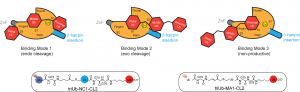
Catalysis and inhibition of deubiquitinases (DUBs)
DUBs antagonize the activities of ubiquitin ligases. It has become clear that DUB activities are indispensable for the normal functions of ubiquitin-proteasome system. Abnormal cellular expression of DUBs or the loss of function due to mutation in certain DUB genes have been linked to various human diseases. We have used high throughput screening to identify DUB inhibitors. The DUB inhibitors were developed and tested in cellular and animal models for anti-cancer therapy. We are developing new assay platforms that allow the efficient screening of DUBs for identification of small molecule inhibitors. We also investigate the catalysis, target protein specificity and the binding modes of ubiquitin moieties in the distinct DUB ubiquitin-binding sites.
Molecular mechanism of DNA damage tolerance and mitophagy
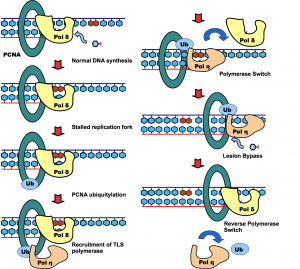 Post-translational modification of proteins represents a crucial way of regulating cellular functions. Modification of cellular proteins by ubiquitin and ubiquitin-like (UBL) proteins plays an essential role in a number of biological processes. New pathways regulated by ubiquitin and UBL are being discovered at a fast pace, virtually in every important aspect of cell biology, including DNA damage tolerance, innate immune response and mitophagy. We are investigating the eukaryotic DNA damage tolerance and mitophagy, in particular the enzymes and protein complexes regulated by post-translational modification by ubiquitin and ubiquitin-like modifiers. Our investigations help to identify new targets and ways of treating human diseases.
Post-translational modification of proteins represents a crucial way of regulating cellular functions. Modification of cellular proteins by ubiquitin and ubiquitin-like (UBL) proteins plays an essential role in a number of biological processes. New pathways regulated by ubiquitin and UBL are being discovered at a fast pace, virtually in every important aspect of cell biology, including DNA damage tolerance, innate immune response and mitophagy. We are investigating the eukaryotic DNA damage tolerance and mitophagy, in particular the enzymes and protein complexes regulated by post-translational modification by ubiquitin and ubiquitin-like modifiers. Our investigations help to identify new targets and ways of treating human diseases.