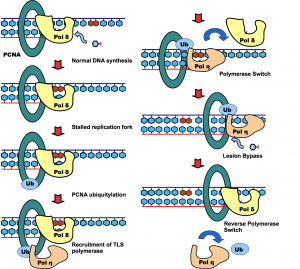
 Posttranslational modification of proteins represents a crucial way of regulating cellular functions. Modification of cellular proteins by ubiquitin and ubiquitin-like proteins plays an essential role in a number of biological processes. Ubiquitin (Ub) modification was initially discovered as a signaling mechanism for proteasome-mediated protein degradation. In recent years, ubiquitin has been found to play far broader roles in eukaryotic cells. New pathways regulated by ubiquitin are being discovered at a fast pace, virtually in almost every important aspect of cell biology, including DNA damage repair/tolerance, signal transduction, transcription, nuclear transport and innate immune response. We are investigating the eukaryotic translesion synthesis (TLS) and its regulation by ubiquitylation and SUMOylation of proliferating cell nuclear antigen (PCNA).
Posttranslational modification of proteins represents a crucial way of regulating cellular functions. Modification of cellular proteins by ubiquitin and ubiquitin-like proteins plays an essential role in a number of biological processes. Ubiquitin (Ub) modification was initially discovered as a signaling mechanism for proteasome-mediated protein degradation. In recent years, ubiquitin has been found to play far broader roles in eukaryotic cells. New pathways regulated by ubiquitin are being discovered at a fast pace, virtually in almost every important aspect of cell biology, including DNA damage repair/tolerance, signal transduction, transcription, nuclear transport and innate immune response. We are investigating the eukaryotic translesion synthesis (TLS) and its regulation by ubiquitylation and SUMOylation of proliferating cell nuclear antigen (PCNA).
We are interested in deciphering the roles of the protein or protein complexes involved in the DNA damage repair/tolerance pathways, in particular the enzymes responsible for the dynamic process of ubiquitylation or SUMOylation, as well as the functional outcomes of the post-translational modification by ubiquitin and ubiquitin-like modifier. We are also trying to discover novel ubiquitin pathways in DNA damage response using DNA microarray, bioinformatics and genetic approaches. Once we identify the target proteins we apply enzymological and structural approaches for the in-depth characterization of the protein with the goal of understanding the catalysis and regulation. Our investigations have important impact on human health, particularly, human cancer. In parallel to our mechanistic studies we are developing the small-molecule and peptidomimetic antagonists that can modulate the essential factors and enzymes in DNA damage response pathway. Some of these molecules will serve as the drug lead for the next generation of anti-cancer therapy.
Reference:
Chen J., Ai Y., Wang J., Haracska J., Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis (2010) Nature Chemical Biology. 6:270
Zhuang Z*, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ*. Regulation of polymerase switch between Polη and Polδ by monoubiquitylation of PCNA and the movement of DNA polymerase holoenzyme. (2008) Proc Natl Acad Sci U S A. 105:5361-5366 (* corresponding author)
Tsutakawa S, Van Wynsberghe A, Freudenthal B, Weinacht C, Gakhar L, Washington M, Zhuang Z, Tainer J, Ivanov I. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. (2011) Proc. Natl. Acad. Sci. USA. 108 (43):17672
Ai Y, Wang J, Johnson RE, Haracska L, Prakash L, Zhuang Z. A novel ubiquitin binding mode in the S. cerevisiae translesion synthesis DNA polymerase η (2011) Molecular BioSystems. 7 (6): 1874
Zhuang Z*, Ai Y. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis (2010) BBA-Proteins and Proteomics. 1804:1081
Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, & Jentsch S, RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. (2002) Nature 419: 135-141.
